Structural biology assignment on Drug Designing
Question
Task: Write a structural biology assignment analyzing the significance of structural biology on drug designing.
Answer
Significance of Structural Biology on Drug Designing: Tackling the effects of mutations in genetic disease and drug resistance
Introduction to the theme of structural biology assignment:
Since the 1980s, for structural biologists designing structured based rational drug design using the structures of protein was an unrealized goal. The first successful project was published in the 90s which was started during the mid-80s. In present there is a scope of improvement to perfect the process, drug designing based on structural biology is an important part of the drug designing programs in most of drug designing programs and is the major research subject of several academic institutions[1]. So, the aim of the literature review on structural biology assignment is to understand how structural biology can be used for drug designing. It will also emphasize on the structure to know the effects of mutation in genetic disorders as well as drug resistance in infectious disease and cancers. Also will emphasize on the possible ways how structural understanding will be advantageous in repurposing off-license rational drugs as well as guide in designing drugs of novel molecules that can be less susceptible to the antibiotic or drug resistance eventually.
In this structural biology assignment, with the accomplishment of the human genome project, the progress in information technology and the beginning of the revolution in both proteomics and structural genomics are powering a much greater opportunity for including designing rational drug based on structural biology in the discovery of the new drugs. By utilizing developments in the field of bioinformatics excellent drug targets are being recognized at a faster pace[2]. The target genes have been quickly cloned and the protein been expressed as well as purified to homogeneity.
Advances in crystallography, like automation at every stage, extra-strong synchrotron radiation, and innovative developments in the determination of phase, have reduced the time period for unfolding the structures. Determination of the structure by means of nuclear magnetic resonance (NMR) has also witnessed several advances in the last few years, comprising both probe and magnet improvements, automatic processing assignment and several new experimental methods to regulate larger structures[3]. The readings provided in the structural biology assignment signifies that advanced computers and their availability at low cost have increased the speed of the drug leads identification process and evaluating it in silico [4].
Structure-based rational drug designing is at its best when it is included in the entire drug discovery process. According to many researchers, structural based designs can lead to a synthesis of compound library based on the combination of structural biology and combinatorial chemistry [2]. Structure-based rational drug design also plays a significant role in the discovery of drug leads (though it is not a drug product it is a compound having a micro-molar affinity towards the target).
The time dedicated to this drug designing processas outlined in this review on structural biology assignment, may signify only a fraction of the entire time taken to develop a saleable drug product. Many more years of research may be needed to transform a drug lead into a product that will be tolerated as well as effective in the human body.
Discovery of Drug based upon structure biology:
The usage of protein structure in the process of drug discovery came up at the same time as the emergence of 3D structures explained through X-ray crystallography[3]. Few of the discoveries that came up are: Cambridge’s Perutz laboratory was the place where the idea behind sickle cell disease was stimulated after observing the hemoglobin’s structure, and the active-site cleft was described through the structure of lysozyme at Philip’s laboratory of London’s Royal Institution in 1966 [3]. Since insulin crystals were used in the treatment of diabetics, insulin’s structure was of particular interest in the pharma industry. The research on structural biology assignment on various sequences of insulin across the globe simulated the thoughts for the design of insulins whose effect lasts longer and is more effective. Further discovery of more sequences of insulin guided the attempts to replicate a homolog of lysozyme present in milk, known as human ?-lactalbumin, and insulin-like growth factor and relaxing during the later stages, which had the biomedicine industry interested in[4].
Other than the laboratories associated with the pharmacy industry, the increment in the use of structure-based drug discovery is observed only after the experimental structure of various enzymes, and their complexes were made available to academic laboratories. This was often done for paralogues which were the industry’s drug target[5].
X-ray crystallography:
The resolution for a frozen conformation’s average structure was found to be in moderate range, i.e. 1.8 Å – 2.5 Å, at the time of determination of protein’s crystal structure. As examined in the structural biology assignment, after the synchrotron radiation facilities came into being, structures with higher resolution, 0.8 Å – 1.2 Å, were observed[5]. Through these structures led to the observations about alternative conformations in few regions of proteins. Even more high resolution structures might be possible in the future, with these radiation facilities being accessible for a larger number of researchers. With the help of co-crystallization, it is possible to uncover information regarding the active site conformations. The process happens in the process of products, intermediates or substrates[6]. This information helps in further analyzing the potential molecules which might be used in drug designing.
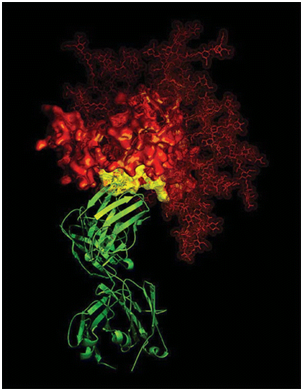
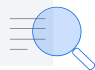
Figure 1: Section of X-ray Crystallography. The image shows a neutralizing antibody in green, B12 with the target in yellow for the vaccine developers in red, HIV-1 gp120. Source: NIAID
Cryo-Electron Microscopy:
According to the research on structural biology assignment, this is extensively used currently in drug discovery, as it helps to better understand pathogenesis and cellular processes. The Zika virus outbreak, which was declared as a public health emergency by WHO, was studied using this technique. A structure of 3.8Å resolution was provided for the virus through the technique, which was further compared to other flaviviruses’ structures in order to determine the antigenicity, pathogenesis and virology of the Zika virus[5]. This analysis illustrated in the structural biology assignment led to the discovery of the region which served as the host to the virus and made the disease more severe.
The technique gives the advantage to a researcher that the macromolecules can be observed in their hydrated state, which is very close to their natural state. Herein structural biology assignment, this ability proves to be critical step towards understanding the relationship between structure and its function[8]. At the end, this technique looks promising in drug discovery by providing structural details of biomolecules which would have been difficult with other structural techniques.
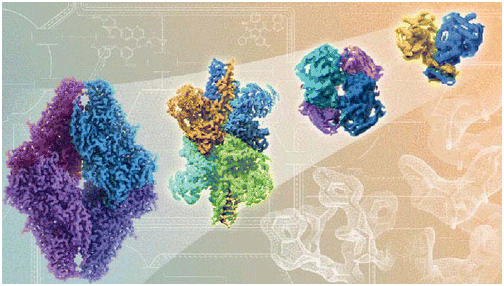
Figure 2: Section of Cryo-electron microscopy. The advancement in the technique is represented in the image with the improvement in resolution of the protein’s atomic details.Source: NIAID
Mutations and Disease:
Many of the mutations related to disease impact the protein functions. These mutations are generally residues that are either varied conservatively or are conserved during evolution. In order to understand the genetic diseases, it becomes important to classify and interpret mutations associated with disease and non-disease [6]. Through the disease-causing mutations’ structural analyses, it was found that most of them happen at the hydrogen-bonded and solvent-inaccessible residues. To calculate the mutation effect on a protein’s stability, a statistical potential change of a residue across its evolution can be used. Site-Directed Mutator (SDM)presented in this section of structural biology assignmentis one of such methods. It makes use of a set of substitution tables that are environment-specific and conformational constrained. These tables consider confirmation of a residue, hydrogen bonding in side-chain, accessibility of solvent, in order to calculate the variation between the stability of a mutant protein structure and a wild-type protein structure. Such methods have led to evidence of the role of mutations in various human genetic diseases like cancer. Consider the case of von Hippel-Lindau Syndrome for example in the present context of structural biology assignment, which is caused by the VHL gene mutations (Figure 1) and leads to development of clear cell renal carcinoma.
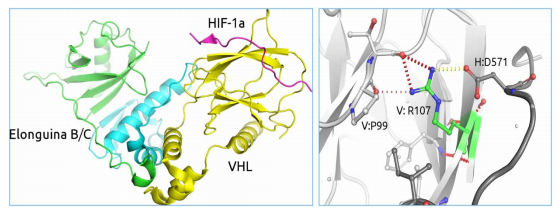
Figure 3[1]: (On the left) In order to identify the risk of a patient to develop clear cell carcinoma in the given scenario of structural biology assignment, the effect of mutations on the VHL protein stability and alteration in their affinity for binding partners (Elongin B and C, and HIF-1?) can be observed.(On the right) Arginine(R107) mediated inter-subunit interactions. The charge complementarity of subunits is altered through mutations in VHL disease, which leads to destabilization of protein-protein interactions.
In other research on the human Mendelian diseases conducted on this structural biology assignment, it was observed that apart from the mutations that affect the stability of protein, there are mutations that affect protein-protein interactions that are important [7]. Remarkably, changes in the stability of the protein have also been related to population phenotypic variability, which includes responses of the drug.
Development of drug resistance is also driven by the same molecular consequences. Strong correlations between various mutations’ structural feature and antibiotic sensitivity link of theirs can be understood by observing the single point coding mutation in Mycobacterium tuberculosis [5]. This theory depicted in the structural biology assignment can help the researchers to predict the resistance mutations prior to their formation, which can in turn help in developing optimal therapeutic interventions and drug development guidance. By applying this theory researcher tried to develop micro molecule inhibitors of GuaB2 for the treatment of Mycobacterium tuberculosis. With the help of VCC234718, analysis of GuaB2 crystal structure revealed that at Y487 the most expected resistance mutation will occur. This will affect the interactions with the inhibitor and alter it but will not alter NAD, and also will not disrupt stability of the protein as well as homo-tetramer unit’s interactions. The major resistance mutation that was observed is Y487C[1]. This resistance mutation can be treated by altering the interactions and inhibitor modification. Molecules other than these that were active and were not part of these interactions were identified (Figure 2). Though through the GuaB2 inhibition, it was observed that the inhibitors were active against Mtb in vitro, further research is needed to find out the therapeutic effects of in vivo inhibition of GuaB2.
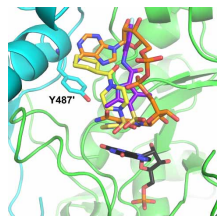
Figure 4[1]:The yellow sticks, GuaB2 on top of black sticks, IMP, which is similar to orange sticks, NAD. But, they have different interactions with promoter unit on the side via Y487. The Compound 6, purple sticks, closely mimics the NAD interactions.
The various research discussed above within this structural biology assignment point out at the importance of a mutation’s structure while trying to understand its biological and molecular consequences [4]. In order to understand longer-range allosteric effects and to differentiate mutations that cause disease, classification based on the solvent accessibility of the residue is not enough. This shortcoming led to a consideration of other structural features in order to locate the residue in the structure’s interiors.
Future prospects of drug discovery based on structure biology: There is clear evidence present from various researches considered in this segment of structural biology assignment about the role of both experimental and computational structure-guided approaches in the early drug discovery. The impact of the mutation on drug resistance and human genetic diseases can also be understood through structures [1]. Numerous researchers have shown that the mechanism of genetic disease caused by mutations, involve disruption in interfacial space and effects that are allosteric on multiprotein assemblies and other functions of the oligomeric protein.
The development of the interfacial stabilizers suggests an important approach that depends on a depth understanding of the structure of protein. Further methods to combat resistance comprise the usage of peptide inhibitors that have way more core flexibility and frequently able to adopt different conformations in order to perform other interactions with the mutant or altered protein[11].
Conclusion: It is concluded from the overall discussion on structural biology assignment that structure-based rational drug designing is a most powerful method, mainly when this method is used within an armamentarium as a tool to discover novel drug leads against pivotal targets. After a primary assessment of bioavailability, the drug leads remain to continue in a repetitive process of reentering in the determination stage of the structure and reevaluation for optimization[12].
According to researchers in order to identify the resistance and determine the nature of various viruses in several infectious diseases, knowing the structural conformations (by utilizing several techniques like NMR, Crystallography and Cryo-Electron Microscope) of the protein targets is very important. In the process of overcoming the resistance, a primary objective is to design a drug that can interact with the residues that are being utilized by the natural lights [5]. It is very important to know the interactions of the drug beyond their site of cofactors, so mutagenesis in silico can be used to determine the ways fragments will grow as well as in redesigning the drug in a way that it will give less resistance mutations. By observing all the possible ways provided in the structural biology assignment, it seems that structural biology is a pivotal component of future research in the way of drug discovery.
References
- D. Sirohi, Z. Chen, L. Sun, T. Klose, T. C. Pierson, M. G. Rossmann and R. J. Kuhn, "The 3.8 Å Resolution cryo-EM Structure of Zika Virus," Science,structural biology assignmentvol. 352, no. 6284, p. 467?470, 2016.
- A. P. Pandurangan, D. B. Ascher, S. E. Thomas and T. L. Blundell, "Biochemical Society Transactions," Genomes, structural biology and drug discovery:combating the impacts of mutations in genetic and antibiotic resistance , vol. 45 , no. 2, p. 303–311, (2017).
- F. Sohraby and H. Aryapour, "Rational drug repurposing for cancer by inclusion of the unbiased molecular dynamics simulation in the structure-based virtual screening approach: Challenges and breakthroughs," Seminars in Cancer Biology, 2020.
- P. Selzer, R. Marhöfer and K. O., "Protein Structures and Structure-Based Rational Drug Design," in Applied Bioinformatics, Springer, Cham, 2018, pp. 73-89.
- R. R. Ramsay and G. D. Giovanni, "Editorial: Structure-Based Drug Design for Diagnosis and Treatment of Neurological Diseases," Frontiers in Pharmacology, vol. 8, p. 13, 2017.
- H. Jubb, A. Pandurangan, M. Turner, B. Ochoa-Montaño, T. Blundell and D. Ascher, "Mutations at protein-protein interfaces: Small changes over big surfaces have large impacts on human health," Prog Biophys Mol Biol, vol. 128, pp. 3-13, 2017.
- J. R. Helliwell, "New Developments in Crystallography: Exploring Its Technology, Methods and Scope in the Molecular Biosciences," Biosci Rep, vol. 37, no. 4, 2017.
- J. Holcomb, N. Spellmon, Y. Zhang, M. Doughan, C. Li and Z. Yang, "Protein Crystallization: Eluding the Bottleneck of X-ray Crystallography," AIMS Biophys, vol. 4, no. 4, pp. 557-575, 2017.
- J. B. Park, Y.-J. Kim, J. M. Yoo, Y. Kim, S.-M. Kim, S. J. Kim, R. Gorbachev, I. I. Barbolina, M.-H. Yoon, B. H. Hong and K. S. Novoselov, "Non-destructive electron microscopy imaging and analysis of biological samples with graphene coating," 2D Materials, vol. 3, no. 4, 2016.
- W.-S. Ryu, " Virus structure," in Molecular Virology of Human Pathogenic Viruses , Boston, Academic Press, 2017, p. 21–29.
- W. J. Zuercher, J. M. Elkins and S. Knapp, "The Intersection of Structural and Chemical Biology-An Essential Synergy," Cell Chem Biol, structural biology assignmentvol. 23, no. 1, p. 173?182, 2016.
- R. F. Thompson, M. Walker, C. A. Siebert, S. P. Muench and N. A. Ranson, "An Introduction to Sample Preparation and Imaging by Cryo-Electron Microscopy for Structural Biology," Methods, vol. 100, p. 3?15, 2016.